Rtp Family Members Induce Functional Expression of Mammalian Odorant Receptors

Hot Spot Mutagenesis Improves the Functional Expression of Unique Mammalian Odorant Receptors
ane
Section of Biotechnology and Life Scientific discipline, Tokyo University of Agriculture and Technology, Tokyo 184-8588, Nippon
2
Instrumentation Assay Center, Tokyo University of Agriculture and Technology, Tokyo 184-8588, Japan
iii
Biological Process of Aging, Tokyo Metropolitan Institute of Gerontology, Tokyo 173-0015, Nihon
iv
Department of Molecular Genetics and Microbiology, Knuckles University Medical Centre, Durham, NC 27710, USA
*
Writer to whom correspondence should be addressed.
Bookish Editors: Masoud Jelokhani-Niaraki and Edgar Soria-Gomez
Received: 19 Nov 2021 / Revised: 15 December 2021 / Accustomed: 24 December 2021 / Published: 28 Dec 2021
Abstract
Vertebrate animals observe odors through olfactory receptors (ORs), members of the G protein-coupled receptor (GPCR) family. Due to the difficulty in the heterologous expression of ORs, studies of their odor molecule recognition mechanisms have progressed poorly. Functional expression of most ORs in heterologous cells requires the co-expression of their chaperone proteins, receptor transporting proteins (RTPs). Yet, some ORs were found to be functionally expressed without the support of RTP (RTP-independent ORs). In this study, nosotros investigated whether amino acid residues highly conserved amongst RTP-contained ORs improve the functional expression of ORs in heterologous cells. We found that a single amino acid commutation at one of two sites (NBWiii.39 and 3.43) in their conserved residues (Eastward and 50, respectively) significantly improved the functional expression of ORs in heterologous cells. Due eastiii.39 and Lthree.43 as well enhanced the membrane expression of RTP-dependent ORs in the absenteeism of RTP. These changes did non alter the odorant responsiveness of the tested ORs. Our results showed that specific sites within transmembrane domains regulate the membrane expression of some ORs.
one. Introduction
Olfaction is an essential awareness for animals to keep carrying out their daily life activities while experiencing changes in the surrounding environment. Mammalian olfactory receptors (ORs) are expressed on the prison cell surface of olfactory sensory neurons (OSNs) to detect and discriminate the vast number of odors in the environment [one]. ORs form the largest and well-nigh various family of G protein-coupled receptors (GPCRs) [2]. There are more than 400 and 1200 functional ORs in the olfactory sensory neurons of humans and mice, respectively [3,4,5]. Information technology is thought that this diverse odor recognition of ORs has made it possible for olfaction to discriminate odors, which are a mixture of various volatile molecules, with loftier sensitivity [six,7]. A major difficulty in characterizing the part of these mammalian ORs is that well-nigh ORs are not exported to the plasma membrane and are instead retained in the endoplasmic reticulum (ER) in nonolfactory cells, making in vitro characterization extremely challenging [2]. Consequently, functional analysis of ORs is far behind that of other canonical Class A GPCRs. Therefore, the flexible and selective scent molecule recognition mechanism of ORs is almost unknown. To fill this gap in knowledge, information technology is necessary to develop a method for improving OR expression without changing the function of ORs. The use of a Rho tag or IL6 tag attached to the North-terminus of ORs has been suggested every bit a method due to the technology's ability to promote the enhancement of surface expression of ORs [viii,9,ten]. Additionally, co-expression of chaperone proteins, Hsp70t or Ric-8B, is constructive for the functional expression of some ORs [xi,12]. Nonetheless, both modifications only enhance the expression of a limited number of ORs.
Heterologous expression of many ORs is achieved with the assistance of receptor transporting protein (RTP) 1S and RTP2 [13,xiv,15,16,17]. These are unmarried transmembrane proteins, a grade of chaperones, that mediate the send of ORs into the plasma membrane. However, the detailed mechanism of the membrane trafficking of ORs by RTPs is unknown. Interestingly, a small subset of ORs can localize to the cell membrane without RTP1 and RTP2 in heterologous cells and function even in double knockout mice lacking RTP1S and RTP2 [xviii]. We previously demonstrated the importance of a specific amino acid residue within transmembrane domain 4 (Chiliad4.53) for the functional expression of OR in heterologous cells. Poor cell surface expression seems to correlate with structural instability [xix].
This study investigated whether mammalian OR expression could be improved by replacing an amino acid with one that is highly conserved in a well-expressed OR (RTP-independent OR). We also tried to identify of import amino acrid residues that contribute to the functional expression of mammalian ORs.
2. Results
ii.i. Screening of the Amino Acid Mutations That Enhance OR Expression
We hypothesized that highly conserved amino acid residues in RTP-independent ORs play essential roles in OR trafficking in heterologous cells. Our previous study showed that amino acid residues at 66 sites are associated with RTP independence [19]. Amid them, the conserved amino acrid residues at 26 sites appeared more often in RTP-independent ORs than in all intact 1093 mouse ORs. Their positions were dispersed throughout the OR sequence (Figure 1A and Supplementary Data S1). We hypothesized that these ORs that accept non-conserved amino acrid residues at these positions might show higher levels of expression when these residues are changed to conserved ones.
We tested this hypothesis past constructing 40 mutant ORs, transiently expressing them in HEK293T cells and analyzing their cell surface expression past FACS. We commencement tested fourteen sites out of the 26 in 14 RTP-independent ORs (Figure 1A Red circumvolve). Among them, Olfr544 (also known equally OR-S6), D115E (NBW3.39), Olfr194_128H (IC2), and Olfr78_Q297E (C-term) exhibited a significant increase in surface expression (Figure 1B, Figures S1 and S2).
Nosotros adjacent tested whether the changes to the conserved residues in these sites improve the expression of RTP-dependent ORs. In the same mode, we changed a non-conserved amino acid residue to a conserved remainder in 11 RTP-dependent ORs. Ninety-seven single amino acid mutants covering all 26 sites were synthetic and expressed in HEK293T cells without RTP1S coexpression. Ii mutants (Olfr992D111E (NBWiii.39) and Olfr982Y120L (NBW3.43)) showed enhanced plasma membrane localization by more than than xxx% (Figure 1C, Figures S1 and S3). Since NorthwardBWiii.39 and NBWiii.43 are in proximity in the same transmembrane helix, interactions betwixt them may contribute to the expression of ORs. Based on these screenings with both RTP-dependent and RTP-independent ORs, we decided to focus on our study on residues within transmembrane domain 3.
two.2. Improvement of Surface Expression by the 3.39E and three.43L Mutations in Many ORs
In 1093 intact mouse ORs registered in the HORDE database [21], E(glutamic acid) accounted for 85.iv% (933/1093) of NBW 3.39, and Fifty(leucine) accounted for 93.3% (1020/1093) of NBW three.43 (Figure 2A,B). To examine whether mutations of nonconserved NBWthree.39 and NBW 3.43 residues to E (iii.39E mutation) and L (iii.43L mutation) are effective for expressing various ORs, nosotros applied the mutations to mouse ORs with known ligands. We fabricated iii.39E mutants for six ORs (Olfr339, Olfr992, Olfr1352, Olfr1353, Olfr1377, and Olfr960) and 3.43L mutants for v ORs (Olfr62, Olfr982, Olfr978, Ofr979, and Olfr960) and measured the mutant expression level on the cell surface (Figure 2A–C). Surface expression on heterologous cells was improved for all single mutants except Olfe1352 D3.39E in the co-expression of RTP1S. Furthermore, half-dozen ORs were localized on cell membranes without co-expression of RTP1S later introducing mutations of iii.39E or 3.43L (Figure second). The cell surface expression of Olfr960, which has both L3.39 and Y3.43, was not improved by either mutation (L3.39E and/or Y3.43L). However, the effect of RTP1S co-expression on the mutants was higher than that on the wild type (Figure 2C,D)
ii.iii. Olfr544 D3.39E Showed Significantly High Expression without a Change in Ligand Selectivity
The unmarried amino acid mutation D3.39E significantly improved the membrane expression of Olfr544. The expression level of Olfr544D115EBW3.39 alone was significantly higher than with the co-expression of RTP1S (Effigy 3A,B). Olfr544 responds to dicarboxylic acids, including nonanedioic acid (azelaic acrid) [22]. Nosotros tested Olfr544 WT and D3.39E mutant responses to dicarboxylic acids with various carbon chain lengths (Figure 3C). The responses of the D3.39E mutant were higher than those of the WT, correlating with the higher expression level in the mutant. The selectivity for dicarboxylic acids with dissimilar carbon chains was non affected by the mutation. The D3.39E mutant responded to concentrations of nonanedioic acid 100 times lower than the WT, with statistical significance (Figure 3D). The ligand selectivity did not alter for the tested ligands.
Olfr544 belongs to the Course I OR family, and D3.39 is widely conserved in Olfr544 homologs (Effigy S4), suggesting that Olfr544D3.39 appeared relatively early on and was subsequently maintained during evolution. When D3.39 of Olfr544 was mutated to various amino acids, the D3.39E mutant showed the highest cell surface expression level amid the mutants (Effigy S5). The D3.39R and D3.39N mutants showed increased expression simply lost responsiveness to their agonist. Since NorthwardBWiii.39 of ORs is located in the sodium ion bounden site in Course A GPCRs [23,24], it is plausible that the interaction of glutamate with sodium ions is important for the stability and role of ORs.
To gain insights into the mechanism of the comeback of stability by the NBWiii.39E mutation, we constructed structural models of Olfr544 variants, wild type, D3.39E (D115E) mutants, and D3.39R (D115R) mutants using AlphaFold 2 [25]. The overall structures of the models were almost identical. The distance betwixt the side chain of N7.49 and the side chain of D3.39 (D115) is 4.five Å in the wild type. In the D3.39E mutant, the distance between E3.39 and N7.49 was shortened to three.4 Å to form a hydrogen bond (Figure 3E).
In the model of the D3.39R mutant, the arginine balance of NBW3.39 was located in the heart of the lumen of the GPCR, similar to other reported Form A GPCRs (Figure S5) [26]. The effects of the D3.39E and D3.39R mutations are related to the coordination and allosteric action of the sodium ion. In other words, this suggests that NBW3.39 of ORs also functions in the allosteric movement of ORs as a sodium ion binding site, which is similar to other Grade A GPCRs.
ii.4. Agonist Response of NBW3.39E and Due northBW3.43L Mutant ORs
Then, we examined the furnishings of the Due northBW3.39E or NBWthree.43L mutation on the ligand responses of diverse ORs. Near OR mutants showed an improved odour response compared to WT (Effigy 4A–C and Supplementary Data S2), correlating with the increase in surface expression. However, the Olfr992D3.39E and Olfr62F3.43L mutants did not answer to the original ligands of octanoic acid and two-coumaranon, respectively (Figure 4D). Since at that place is a correlation betwixt amounts of OR expression and ligand responsiveness in heterologous cells [17], these results suggest that the mutation sites Olfr992D3.39 and Olfr62F3.43 are likely to play a role in ligand binding.
2.5. The Change in Conserved E3.39 and L3.43 Caused a Loss of Function
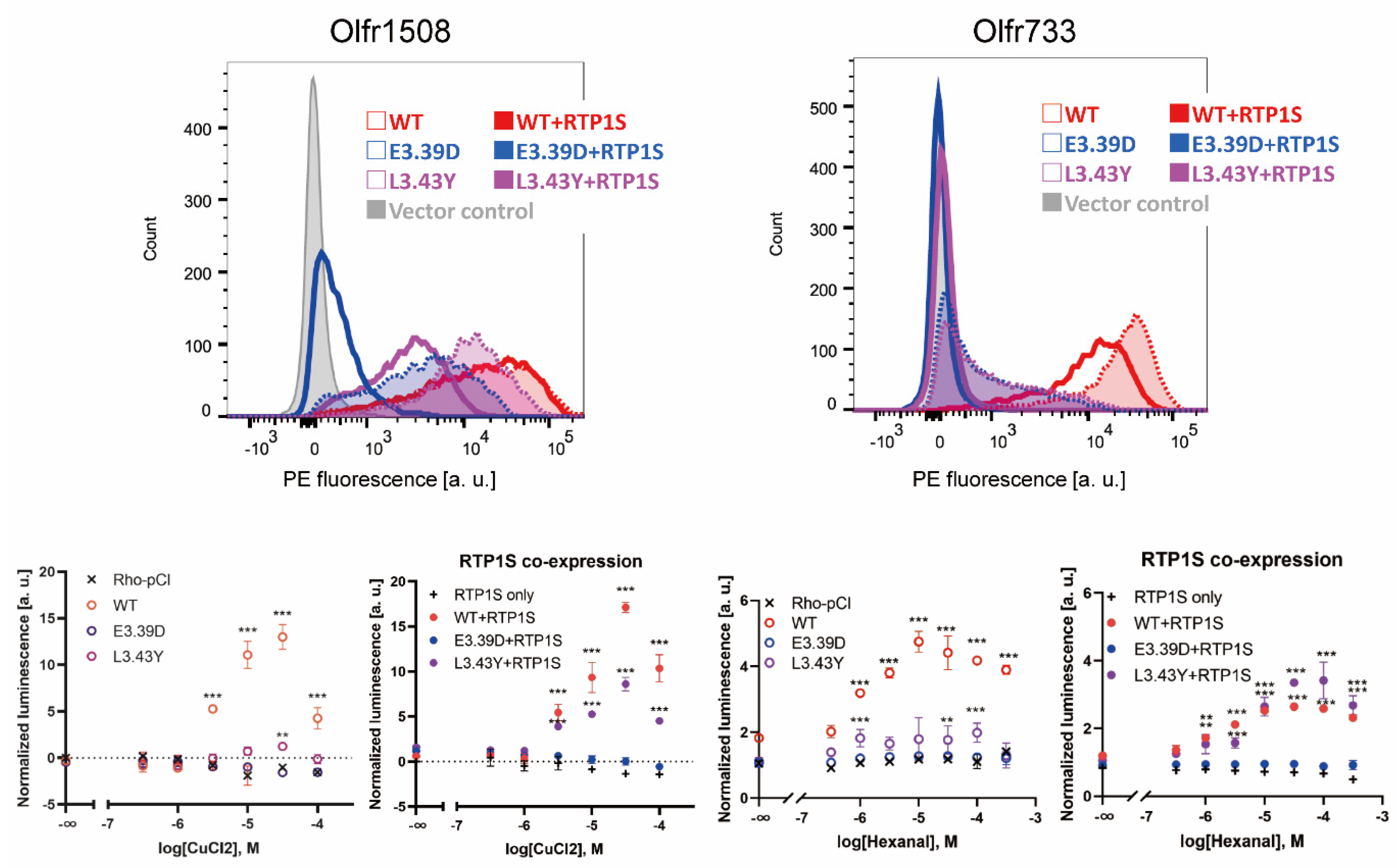
To investigate whether mutation of the conserved NBWiii.39E and Due northBW3.43L alters the functional expression of ORs, we constructed E3.39D and L3.43Y mutants. We selected Olfr1508 (agonist: Cu2+ ion) and Olfr733 (agonist: hexanal), which are highly expressed in the cell membrane without the assistance of RTPs [19,27]. The E3.39D and L3.43Y mutations significantly reduced their jail cell surface expression and ligand response ability (Figure 5).
Although co-expression of RTP1S could rescue the functional expression of L3.43Y mutants, the E3.39D mutant was not recovered. These results suggest that both NBW3.39 and NBWiii.43 of ORs are important residues for the appropriate expression of Ors.
two.half dozen. NBW 3.39E and NBW three.43L Mutations Are as well Effective in Human Ors
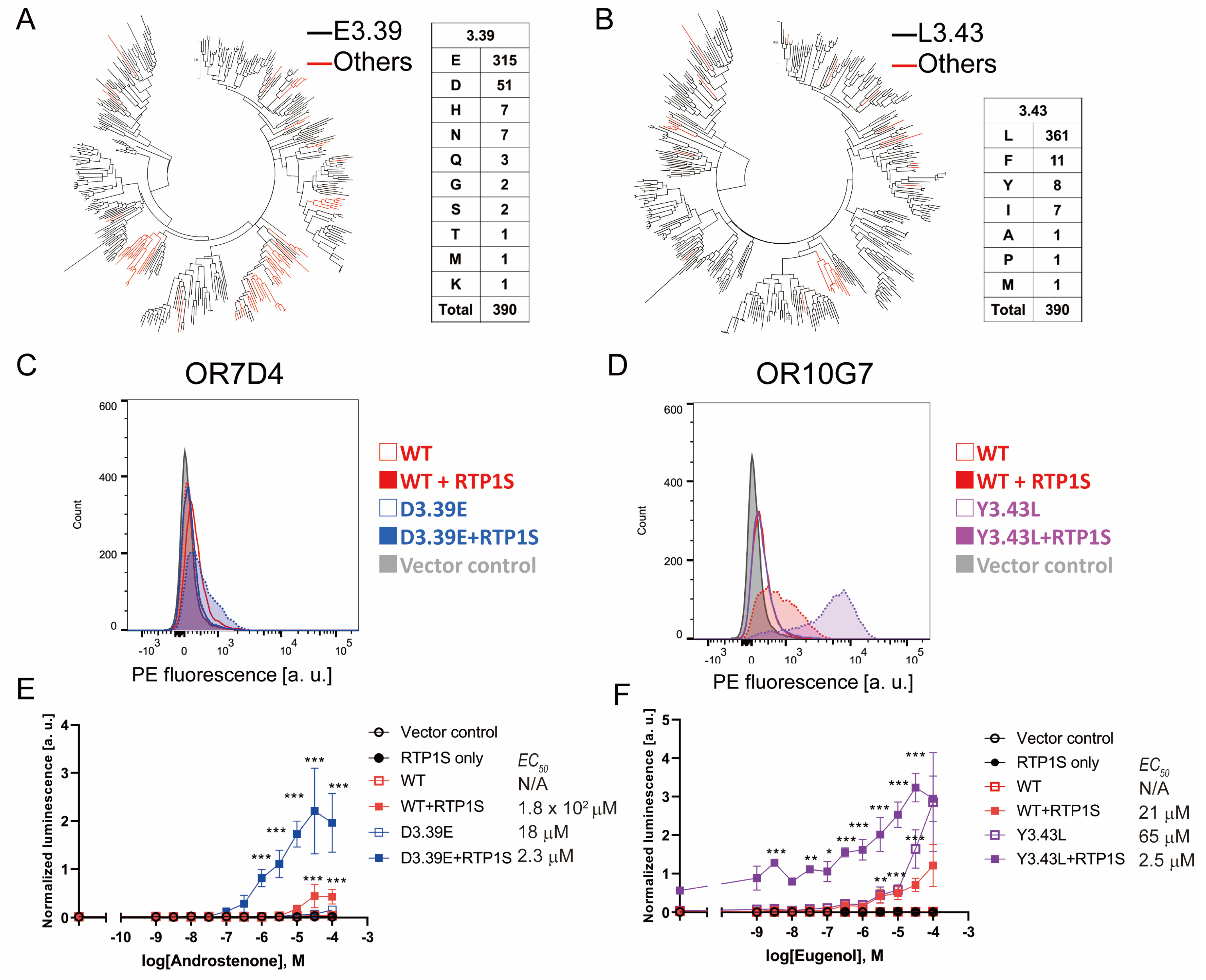
We examined the effects of the 3.39E and iii.43L mutations on human Ors. In human Ors, E(Glutaminc acrid) accounted for 81% (315/390) of NBw3.39 and Fifty(Leucine) accounted for 92.5% (361/390) of NorthwardBw3.43. (Effigy 6A,B). Ors without conserved E3.39 tend to be clustered in a few families, suggesting evolutionary advantages in these families. On the other hand, the Ors whose 3.43 was not L were dispersed throughout the phylogenetic tree, and many of them were pseudogenes. Then, we synthetic iii.39E and 3.43L mutants and examined their surface expression and ligand response (Figure half-dozen and Figure S6). OR7D4 is ane of the characteristic Ors in which human being perception of androstenone changes dramatically depending on genetic variants [28]. The D3.39E mutation significantly increased the cell surface expression of OR7D4, and the detection limit of androstenone (5α-androst-xvi-en-3-1) was improved to less than 1 µM (Figure 6C). A meaning increment was also observed in OR10G7 Y3.43L, which was expressed without co-expression of RTP1S (Figure 6D). This mutant also detected a significant response against Eugenol even at lower stimulus concentrations than the wild type [29]. Mutations in the NBW3.39 and 3.43 amino acids tin can be constructive ways to improve the functional expression of human ORs. Taken together, the amino acids located at NorthBW3.39 and 3.43 are involved in regulating the expression of mammalian ORs.
3. Discussion
Animals can detect various odorants in the environment through blueprint recognition of the response by hundreds of ORs [seven,30]. Nosotros presented a hypothesis that olfactory neurons acquired a loftier level of evolutionally capacitance to allow rapid evolution of ORs [nineteen]. This rapid evolution allows ORs to respond to structurally various environmental odour molecules but compromises conformational stability, resulting in difficulties in functional expression of ORs in not-olfactory neurons [nineteen]. In this study, from the amino acid residues conserved amidst the RTP-independent ORs with high conformational stability, we found that NBW3.39 and 3.43 in the 3rd transmembrane domain are related to OR expression. The iii.39E and iii.43L substitutions were effective in improving the membrane localization of multiple ORs in mice and humans. Curiously, conservation of NBWiii.39E and NBW3.43L in man ORs is lower than that in mouse ORs. This may, in part, explain why expression levels of the human ORs are generally lower than those of mouse ORs in heterologous cell expression systems. 3.39E mutations of the ORs containing D at Due northBW3.39 often enhanced membrane expression (7/10 in tested ORs), while the 3.39E mutation was less effective for ORs with other amino acids at NorthwardBW3.39, such every bit Q (1/4 in tested ORs). Both NBW3.39 and 3.43 are in the transmembrane helices and confront the same direction. Interestingly, Due northBW4.53, which affects the RTP dependency of Olfr539, too faces towards NBW3.39 and NBW3.43 [nineteen]. Together, the luminal surround of each transmembrane helix may be important for the stability of ORs and their associated membrane localization. This may occur via the stabilization of membrane packing. Residues in both TM3 and TM4 of ORs may human activity in combination to stabilize membrane packing. This work successfully identified two specific residues to facilitate the expression of ORs. The expression levels of the human ORs were lower than those of mouse ORs in heterologous jail cell expression systems. By introducing these mutations, detailed analysis of orphan ORs, which could not be analyzed due to its low expression level, may be achieved.
We previously report that the introduction of arginine to specific residues predicted to improve thermal stability also improved the functional expression of consensus ORs [xix]. It has also been reported that the functional expression of OR was improved by introducing mutations to transmembrane domains other than TM3 and TM4 and the C-concluding region [31,32]. These methods that accept been effective to some ORs appears to be ineffective with other ORs. Equally a consequence, no single method can exist adapted to ameliorate the expression of all mammalian ORs. In that location may be dissimilar solutions to the OR trafficking problem depending on the private ORs. Nonetheless, the solutions can exist broadly categorized into a pocket-sized number of methods, ane of which would be to stabilize membrane packing past calculation advisable mutations.
NBW3.39 and NBWthree.43 are located in the vicinity of the sodium ion binding site in other Class A GPCRs. NBWthree.39 is frequently mutated to increase the product yield for crystallization of GPCRs [26,33]. The 8 amino acids out of the 15 sodium-binding sites reported are different from those of ORs to the canonical Course A GPCR (Figure S7), and the relationship betwixt the functional activity of the ORs and the sodium ion has not been reported thus far. This departure in amino acid conservation of the sodium ion-binding site may hateful that the origin of the mammalian ORs is fundamentally different from other GPCRs.
It is very likely that sodium ions are coordinated in the vicinity of Due northBW3.39 and deed allosterically, the same equally in other GPCRs. While the three-dimensional structures of many Class A GPCRs have been reported, none of the ORs have been solved. Reports of ligand docking assay and MD simulation of ORs using a model construction are increasing [34,35,36,37]. The progress of structure prediction technologies including AlphaFold 2 has been remarkable [25]. Our report identifying important residues that contribute to the functional expression of ORs volition help lead to the elucidation of the functional mechanism of ORs and the evolution of high-sensitivity scent sensing engineering using mammalian ORs.
The introduction of the 3.39E or iii.43 Fifty mutation significantly affected the functional expression of ORs. Most mutant ORs retained odorant selectivity except for Olfr544 D3.39R, which acquired a loss of ligand response while improving membrane localization. Although the mutations (NBW3.39E and NBW3.43L) we found in this written report are applicable to a pocket-sized number of ORs (approximately 19% and 7.five% of human being ORs, respectively), the ORs may have evolved to have unique characteristics in odor detection in some cases. Future experiments can address this hypothesis by expanding the number of ORs and odorants tested using NBW3.39E and NBW3.43L mutations. Currently, withal, the bulk of mammalian ORs are notwithstanding poorly expressed even with the help of RTPs, suggesting that pieces are nevertheless missing for the stable expression of ORs in heterologous cells.
4. Materials and Methods
iv.1. DNA and Vector Preparation
Open reading frames of OR genes were subcloned into pCI (Promega, Madison, WI, USA) with a rhodopsin tag at the N-terminus. To generate mutants of ORs, DNA fragments of OR genes were amplified by Phusion polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and PrimeStar MAX polymerase (Takara bio, Shiga, Nihon). The fragments were mixed and amplified by PCR to obtain full sequences. All plasmid DNA was purified past NucleoSpin plasmid transfection grade (MACHEREY-NAGEL GmbH & Co, Düren, Germany). Other expression vectors were the same as used in a previous study [17]. All plasmid sequences were verified using Sanger sequencing.
iv.2. Cell Culture
HEK293T and Hana 3A [xiii] cells were grown in Minimal Essential Medium (MEM) containing 10% FBS (vol/vol) with penicillin-streptomycin and amphotericin B. Hana 3A cells were authenticated using polymorphic short tandem repeats (STRs) at the Duke Deoxyribonucleic acid Analysis Facility using GenePrint ten (Promega) and shown to share profiles with the reference (ATCC). All prison cell lines were incubated at 37 °C, saturating humidity and five% COtwo. No mycoplasma infection was detected in any jail cell culture.
4.3. Menstruation Cytometry Analyses
HEK293T cells were grown to confluence, resuspended, and seeded onto 35 mm plates at 25% confluency. The cells were cultured overnight. The Rho-tagged OR in the plasmid pCI and GFP expression vector were transfected using Viafect transfection reagent (Promega). Afterward 18–24 h, the cells were resuspended in a cell stripper (Corning, NY, Usa) and then kept on ice. The cells were spun down at 4 °C and resuspended in PBS containing fifteen mM NaNthree and 2% FBS to wash away the jail cell stripper. They were then incubated with a chief antibody mouse anti-Rho4D2 mouse antibody (Merck-Millipore, Burlington, MA, U.s.a.), washed, and so stained with phycoerythrin (PE)-conjugated donkey anti-mouse F(ab')₂ fragment antibody (Abcam, Cambridge, UK) in the dark. To stain expressionless cells, seven-amino-actinomycin D (Merck-Millipore, Burlington, MA, USA) was added. The cells were analyzed using BD FACSCanto Two FACS and BD LSRFortessa with gating allowing for GFP-positive, single, spherical, viable cells, and the measured PE fluorescence intensities were analyzed and visualized using FlowJo.
4.iv. Immunocytochemistry
Live-jail cell surface staining was performed as described previously [38]. The primary antibody used was mouse anti-rhodopsin 4D2 (Merck-Millipore). The secondary antibodies used were Cy3-conjugated anti-mouse IgG antibody (Thermo Fisher Scientific, Waltham, MA, United states of america). After antibody staining, the cells were fixed in 4% paraformaldehyde, then the slides were mounted with Mowiol and visualized by a fluorescence microscopy M1 imager (Carl Zeiss, Oberkochen, Deutschland).
four.5. Dual Luciferase Reporter Gene Assay
All odorants were purchased from FUJIFILM Wako Chemicals (Osaka, Japan) and TCI chemicals (Tokyo, Japan). The Dual-Glo Luciferase Assay (Promega) was used to make up one's mind the activities of firefly and Renilla luciferase in Hana3A cells equally previously described [38]. Briefly, firefly luciferase, driven past a army camp response element promoter (CRE-Luc; Stratagene), was used to measure out the OR activation levels. For each well of a 96-well plate, v ng SV40-RL, 10 ng CRE-Luc, five ng mouse RTP1, 2.5 ng M3 receptor 3, and v ng of Rho-tagged receptor plasmid DNA were transfected. Normalized activity for each well was further calculated as (Luc)/(Rluc). Luc and Rluc are the brilliance of firefly luciferase and Renilla luminescence, respectively. The basal activity was averaged from 6 wells in the absence of odorants and farther corrected by subtracting that of the control empty vector. Odorant-induced activity was averaged from at least 3 wells and further corrected by subtracting the basal activeness of that receptor. Odorant-induced responses were normalized to that of WT. EC50 value was calculated using GraphPad Prism software and data shown in Supplementary Data S2.
four.6. Statistical Assay
Multiple comparisons were performed using 1-fashion or two-way analysis of variance (ANOVA) using GraphPad Prism. Student's t tests were performed using the congenital-in function in Microsoft Excel. The boilerplate is shown as the hateful ± standard errors.
Supplementary Materials
Author Contributions
Conceptualization, Y.F. and H.M.; methodology, Y.F.; validation, Y.F.: formal analysis, Y.F.; investigation, Y.F., Y.N., Northward.K., S.M., R.Thousand. and One thousand.I.; resource, M.N. and I.O.; information curation, K.I.; writing—original draft preparation, Y.F., H.Thou. and M.Y.; writing—review and editing, Y.F., H.M. and G.Y.; visualization, Y.F.; supervision, Y.F. and G.Y.; projection administration, Y.F.; funding acquisition, Y.F. and M.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from JSPS-KAKENHI (18K14060 and 20K15745 to Y.F.; 20H02532 to M.Y.) and JST, Human activity-X Grant Number JPMJAX201C to Y.F.
Acknowledgments
We thank Priya Meesa at Duke University for reading and editing the manuscript. YF stayed at Duke University with fiscal support from the JSPS Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers (R2801). KI stayed at Duke University with fiscal support from Tokyo University of Agriculture and Technology as a student of the program for leading graduate schools in Nippon.
Conflicts of Interest
The authors declare no disharmonize of interest.
Abbreviations
| OR | Odorant receptor |
| GPCR | G-protein coupled receptor |
| RTP | Receptor transporting protein |
| BW | Ballesteros-Weinstein number [twenty] |
| ER | endoplasmic reticulum |
| HEK293T | Homo Embryonic Kidney 293T |
| FACS | fluorescence-activated jail cell sorting |
References
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for aroma recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Lu, Yard.; Echeverri, F.; Moyer, B.D. Endoplasmic reticulum retentiveness, degradation, and aggregation of olfactory K-protein coupled receptors. Traffic 2003, iv, 416–433. [Google Scholar] [CrossRef] [PubMed]
- Malnic, B.; Godfrey, P.A.; Buck, L.B. The human olfactory receptor cistron family. Proc. Natl. Acad. Sci. United states 2004, 101, 2584–2589. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, 50.R.; Kondoh, K.; Ye, X.; Yoon, One thousand.H.; Hernandez, M.; Buck, 50.B. Combinatorial effects of odorants on mouse behavior. Proc. Natl. Acad. Sci. USA 2016, 113, E3300–E3306. [Google Scholar] [CrossRef] [PubMed]
- Barnes, I.H.A.; Ibarra-Soria, X.; Fitzgerald, Due south.; Gonzalez, J.M.; Davidson, C.; Hardy, M.P.; Manthravadi, D.; Van Gerven, L.; Jorissen, K.; Zeng, Z.; et al. Expert curation of the human and mouse olfactory receptor gene repertoires identifies conserved coding regions split across ii exons. BMC Genom. 2020, 21, 196. [Google Scholar] [CrossRef] [PubMed]
- Su, C.Y.; Menuz, K.; Carlson, J.R. Olfactory perception: Receptors, cells, and circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef]
- Kida, H.; Fukutani, Y.; Mainland, J.D.; de March, C.A.; Vihani, A.; Li, Y.R.; Chi, Q.; Toyama, A.; Liu, 50.; Kameda, M.; et al. Vapor detection and bigotry with a console of odorant receptors. Nat. Commun. 2018, 9, 4556. [Google Scholar] [CrossRef]
- Krautwurst, D.; Yau, Thousand.Due west.; Reed, R.R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 1998, 95, 917–926. [Google Scholar] [CrossRef]
- Noe, F.; Geithe, C.; Fiedler, J.; Krautwurst, D. A bi-functional IL-6-HaloTag((R)) every bit a tool to measure the jail cell-surface expression of recombinant odorant receptors and to facilitate their action quantification. J. Biol. Methods 2017, iv, e82. [Google Scholar] [CrossRef]
- Noe, F.; Frey, T.; Fiedler, J.; Geithe, C.; Nowak, B.; Krautwurst, D. IL-six-HaloTag((R)) enables live-cell plasma membrane staining, menses cytometry, functional expression, and de-orphaning of recombinant odorant receptors. J. Biol. Methods 2017, 4, e81. [Google Scholar] [CrossRef]
- Neuhaus, E.K.; Mashukova, A.; Zhang, W.; Barbour, J.; Hatt, H. A specific oestrus stupor poly peptide enhances the expression of mammalian olfactory receptor proteins. Chem. Senses 2006, 31, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Von Dannecker, 50.E.; Mercadante, A.F.; Malnic, B. Ric-8B promotes functional expression of odorant receptors. Proc. Natl. Acad. Sci. USA 2006, 103, 9310–9314. [Google Scholar] [CrossRef]
- Saito, H.; Kubota, M.; Roberts, R.West.; Chi, Q.; Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Prison cell 2004, 119, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Matsunami, H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J. Biol. Chem. 2007, 282, 15284–15293. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Pan, Y.; Chen, Yard.Q.; Matsunami, H.; Zhuang, H. Receptor-transporting protein 1 brusk (RTP1S) mediates translocation and activation of odorant receptors past acting through multiple steps. J. Biol. Chem. 2012, 287, 22287–22294. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Su, X.; Pan, Y.; Zhuang, H. Receptor-transporting poly peptide (RTP) family unit members play divergent roles in the functional expression of odorant receptors. PLoS ONE 2017, 12, e0179067. [Google Scholar] [CrossRef]
- Fukutani, Y.; Tamaki, R.; Inoue, R.; Koshizawa, T.; Sakashita, S.; Ikegami, K.; Ohsawa, I.; Matsunami, H.; Yohda, M. The N-terminal region of RTP1S plays of import roles in dimer formation and odorant receptor-trafficking. J. Biol. Chem. 2019, 294, 14661–14673. [Google Scholar] [CrossRef]
- Sharma, R.; Ishimaru, Y.; Davison, I.; Ikegami, Yard.; Chien, Chiliad.South.; Y'all, H.; Chi, Q.; Kubota, Thousand.; Yohda, M.; Ehlers, M.; et al. Olfactory receptor accessory proteins play crucial roles in receptor part and gene choice. Elife 2017, 6, e21895. [Google Scholar] [CrossRef]
- Ikegami, G.; de March, C.A.; Nagai, 1000.H.; Ghosh, S.; Do, M.; Sharma, R.; Bruguera, East.South.; Lu, Y.E.; Fukutani, Y.; Vaidehi, N.; et al. Structural instability and deviation from conserved residues underlie intracellular memory of mammalian odorant receptors. Proc. Natl. Acad. Sci. Us 2020, 117, 2957–2967. [Google Scholar] [CrossRef]
- Juan, A.; Ballesteros, H.W. Integrated methods for the construction of iii-dimensional models and computational probing of construction-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Olender, T.; Nativ, Northward.; Lancet, D. HORDE: Comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013, 1003, 23–38. [Google Scholar] [PubMed]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Jail cell 1999, 96, 713–723. [Google Scholar] [CrossRef]
- Gutierrez-de-Teran, H.; Massink, A.; Rodriguez, D.; Liu, Westward.; Han, Grand.W.; Joseph, J.S.; Katritch, I.; Heitman, L.H.; Xia, L.; Ijzerman, A.P.; et al. The role of a sodium ion binding site in the allosteric modulation of the A(2A) adenosine G protein-coupled receptor. Structure 2013, 21, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Katritch, V.; Fenalti, G.; Abola, E.E.; Roth, B.L.; Cherezov, V.; Stevens, R.C. Allosteric sodium in course A GPCR signaling. Trends Biochem. Sci. 2014, 39, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the homo proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Yasuda, South.; Kajiwara, Y.; Toyoda, Y.; Morimoto, K.; Suno, R.; Iwata, S.; Kobayashi, T.; Murata, T.; Kinoshita, M. Hot-Spot Residues to be Mutated Mutual in One thousand Protein-Coupled Receptors of Class A: Identification of Thermostabilizing Mutations Followed by Conclusion of 3-Dimensional Structures for Two Example Receptors. J. Phys. Chem. B 2017, 121, 6341–6350. [Google Scholar] [CrossRef]
- Li, S.; Ahmed, L.; Zhang, R.; Pan, Y.; Matsunami, H.; Burger, J.Fifty.; Block, E.; Batista, V.S.; Zhuang, H. Smelling Sulfur: Copper and Argent Regulate the Response of Man Odorant Receptor OR2T11 to Low-Molecular-Weight Thiols. J. Am. Chem. Soc. 2016, 138, 13281–13288. [Google Scholar] [CrossRef]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef]
- Mainland, J.D.; Li, Y.R.; Zhou, T.; Liu, W.50.; Matsunami, H. Human olfactory receptor responses to odorants. Sci. Data 2015, ii, 150002. [Google Scholar] [CrossRef]
- Saito, H.; Chi, Q.; Zhuang, H.; Matsunami, H.; Mainland, J.D. Odor coding by a Mammalian receptor repertoire. Sci. Signal 2009, 2, ra9. [Google Scholar] [CrossRef]
- Bubnell, J.; Jamet, S.; Tomoiaga, D.; D'Hulst, C.; Krampis, Grand.; Feinstein, P. In Vitro Mutational and Bioinformatics Analysis of the M71 Odorant Receptor and Its Superfamily. PLoS ONE 2015, x, e0141712. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, M.; Bauer, J.; Haag, F.; Krautwurst, D. Conserved C-terminal motifs in odorant receptors instruct their cell surface expression and cAMP signaling. FASEB J. 2021, 35, e21274. [Google Scholar] [CrossRef] [PubMed]
- Liu, Westward.; Chun, E.; Thompson, A.A.; Chubukov, P.; Xu, F.; Katritch, 5.; Han, G.W.; Roth, C.B.; Heitman, L.H.; Ap, I.J.; et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Scientific discipline 2012, 337, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Abaffy, T.; Bain, J.R.; Muehlbauer, M.J.; Spasojevic, I.; Lodha, South.; Bruguera, E.; O'Neal, S.G.; Kim, S.Y.; Matsunami, H. A Testosterone Metabolite 19-Hydroxyandrostenedione Induces Neuroendocrine Trans-Differentiation of Prostate Cancer Cells via an Ectopic Olfactory Receptor. Front. Oncol. 2018, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Bushdid, C.; de March, C.A.; Topin, J.; Do, 1000.; Matsunami, H.; Golebiowski, J. Mammalian class I odorant receptors exhibit a conserved vestibular-binding pocket. Jail cell. Mol. Life Sci. 2019, 76, 995–1004. [Google Scholar] [CrossRef]
- de March, C.A.; Topin, J.; Bruguera, E.; Novikov, Yard.; Ikegami, Grand.; Matsunami, H.; Golebiowski, J. Odorant Receptor 7D4 Activation Dynamics. Angew. Chem. Int. Ed. Engl. 2018, 57, 4554–4558. [Google Scholar] [CrossRef]
- Charlier, 50.; Topin, J.; de March, C.A.; Lai, P.C.; Crasto, C.J.; Golebiowski, J. Molecular modelling of odorant/olfactory receptor complexes. Methods Mol. Biol. 2013, 1003, 53–65. [Google Scholar]
- Zhuang, H.; Matsunami, H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat. Protoc. 2008, 3, 1402–1413. [Google Scholar] [CrossRef]
Effigy 1. Identification of mutation residues enhancing OR expression. (A) Ophidian plot showing the consensus sequence of mouse ORs. Circle: Amino acid site that constructed the mutant. Ruby: tested on both of RTP dependent and independent ORs, Blueish: tested on RTP dependent ORs just. The table shows the mutation sites and BW (Ballesteros-Weinstein) number [20] tested in this study. (B,C) The cell-surface expression of the RTP independent OR (Olfr194, Olfr544 and Olfr644) and RTP dependent ORs (Olfr982 and Olfr992), respectively. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. X-axis: PE fluorescence, Y-centrality: cell number.
Figure 1. Identification of mutation residues enhancing OR expression. (A) Snake plot showing the consensus sequence of mouse ORs. Circle: Amino acid site that synthetic the mutant. Red: tested on both of RTP dependent and independent ORs, Blueish: tested on RTP dependent ORs simply. The table shows the mutation sites and BW (Ballesteros-Weinstein) number [20] tested in this study. (B,C) The cell-surface expression of the RTP independent OR (Olfr194, Olfr544 and Olfr644) and RTP dependent ORs (Olfr982 and Olfr992), respectively. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. X-axis: PE fluorescence, Y-centrality: jail cell number.
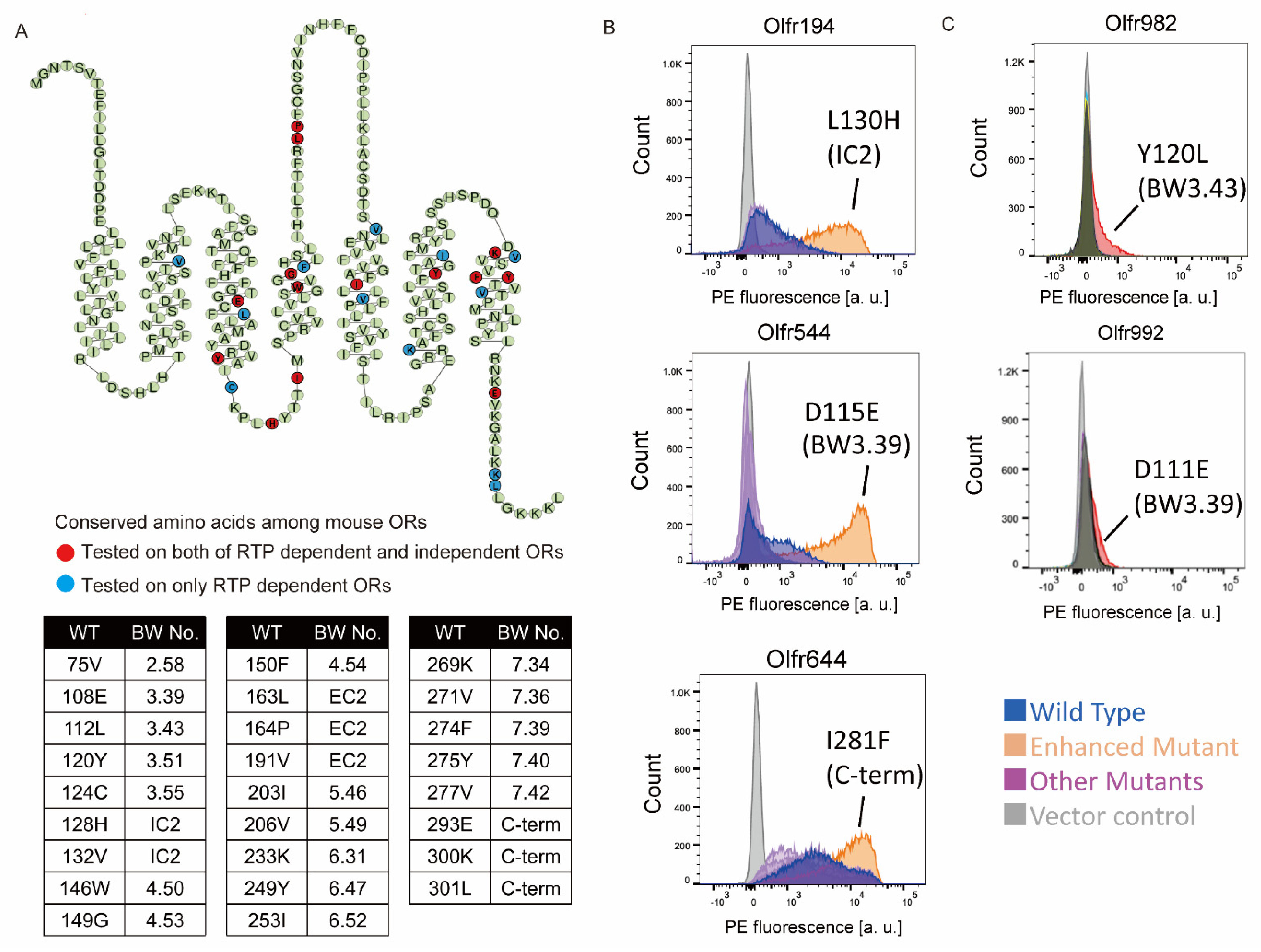
Figure 2. Effect of the 3.39E and 3.43L mutants on mouse ORs. (A) Phylogenic tree of mouse ORs showing the E3.39 or others. Ruddy indicates an OR in which the residue of NBW3.39 is not Due east (Glu) in 1093 intact mouse ORs. Jail cell-surface expression of the Rho-tagged ORs and 3.39E mutants. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. 10-axis: PE fluorescence, Y-axis: prison cell number. (B) Phylogenic tree of mouse ORs showing the L3.43 or others. Carmine indicates an OR in which the residue of NBW3.43 is non 50 (Leu). Cell-surface expression of the Rho-tagged ORs and iii.43L mutants. (C) Jail cell-surface expression of Rho-tagged Ofr960 and its mutants. (D) The values of the event of mutations of three.39E and iii.43L were used every bit the max intensity (cerise) and wild blazon (white) in each OR, respectively.
Figure ii. Effect of the 3.39E and 3.43L mutants on mouse ORs. (A) Phylogenic tree of mouse ORs showing the E3.39 or others. Red indicates an OR in which the residual of NBW3.39 is not Eastward (Glu) in 1093 intact mouse ORs. Jail cell-surface expression of the Rho-tagged ORs and 3.39E mutants. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. X-axis: PE fluorescence, Y-axis: cell number. (B) Phylogenic tree of mouse ORs showing the L3.43 or others. Cherry indicates an OR in which the rest of NBW3.43 is not Fifty (Leu). Cell-surface expression of the Rho-tagged ORs and 3.43L mutants. (C) Jail cell-surface expression of Rho-tagged Ofr960 and its mutants. (D) The values of the effect of mutations of iii.39E and 3.43L were used every bit the max intensity (cerise) and wild blazon (white) in each OR, respectively.
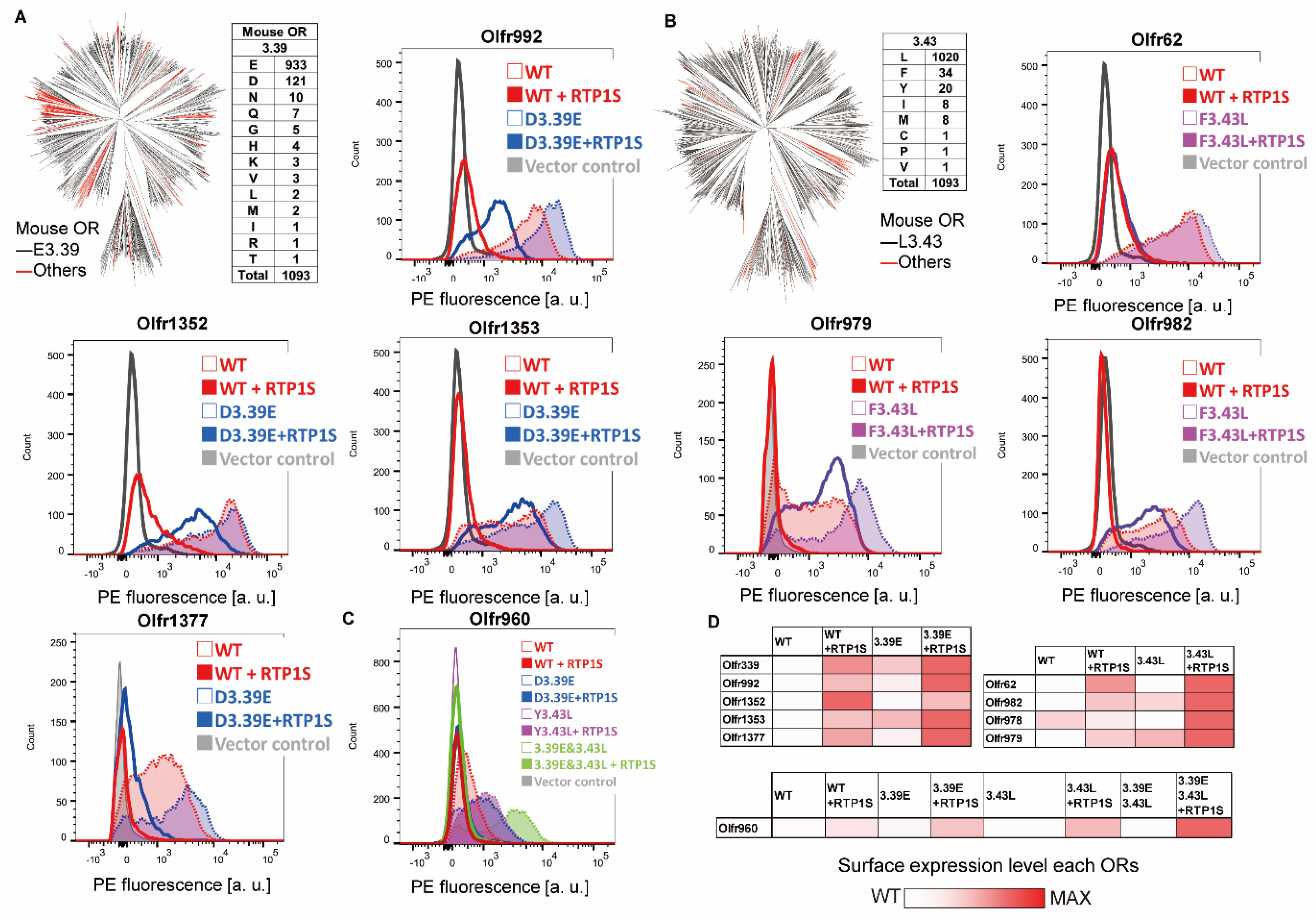
Figure iii. Detailed analysis of Olfr544 D115EBW3.39 (A) Cell-surface expression of the Rho-tagged ORs and D3.39E mutants. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. X-axis: PE fluorescence, Y-axis: cell number. Left: Comparison between the WT and the D3.39E mutant. The results from 2 independent experiments are shown in the graph. Right: The event of RTP1S co-expression of Olfr544 and D3.39E mutant. (B) Immunocytochemical image of Olfr544 and the D3.39E mutant stained with anti-Rho4D2 mouse antibody and PE-conjugated anti-mouse IgG caprine animal antibody. (C) Agonist selectivity of Olfr544 and the D3.39E mutant for various dicarboxylic acids. Error bars point southward.eastward.m (n = 3). (D) Dose response curve of Olfr544 and the D3.39E mutant to nonanedioic acrid. Mistake confined indicate south.e.thousand (n = 3). Multiple comparisons were performed using i-way ANOVA followed by Dunnett's test (* p < 0.05, *** p < 0.001). (E) Structural model of Olfr544 and the D3.39E mutant using Alphafold ii prediction. It was predicted that the distance between the residue of 3.39 and N7.49 was iv.5 Å in the WT (D3.39) and 3.4 Å in the E3.39 mutant.
Figure 3. Detailed analysis of Olfr544 D115EBW3.39 (A) Cell-surface expression of the Rho-tagged ORs and D3.39E mutants. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. Ten-axis: PE fluorescence, Y-axis: cell number. Left: Comparing between the WT and the D3.39E mutant. The results from two independent experiments are shown in the graph. Right: The upshot of RTP1S co-expression of Olfr544 and D3.39E mutant. (B) Immunocytochemical image of Olfr544 and the D3.39E mutant stained with anti-Rho4D2 mouse antibiotic and PE-conjugated anti-mouse IgG goat antibody. (C) Agonist selectivity of Olfr544 and the D3.39E mutant for diverse dicarboxylic acids. Error bars indicate s.east.m (n = iii). (D) Dose response curve of Olfr544 and the D3.39E mutant to nonanedioic acid. Mistake confined indicate s.e.m (north = 3). Multiple comparisons were performed using one-manner ANOVA followed past Dunnett's test (* p < 0.05, *** p < 0.001). (E) Structural model of Olfr544 and the D3.39E mutant using Alphafold ii prediction. Information technology was predicted that the altitude between the residue of iii.39 and N7.49 was 4.5 Å in the WT (D3.39) and iii.4 Å in the E3.39 mutant.
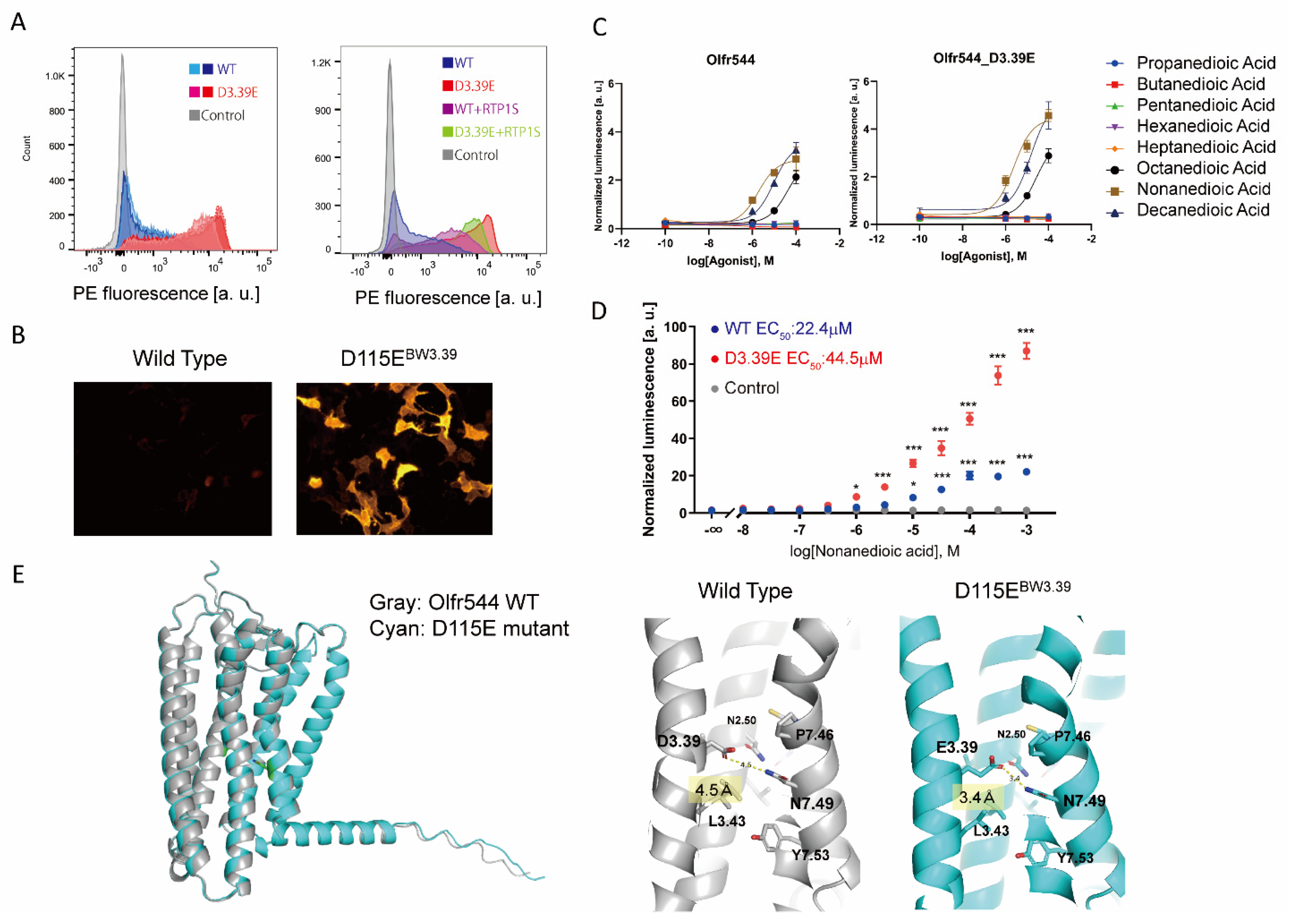
Figure iv. Dose-response curve of each mouse OR mutant. (A) D3.39E mutants of Olfr339, Olfr1352, Olfr1353 and Olfr1377, (B) Y3.43L mutants of Olfr978 and Olfr979, (C) Olfr960 L3.39E, Y3.43L and double mutants, (D) ORs with reduced agonist response due to mutations. Olfr992D3.39E against octanol and Olfr62F3.43L confronting 2-coumaranone. All constructs, including the vector command, were also tested nether RTP1S co-expression conditions. Mistake bars indicate s.e.m (n = three).
Figure 4. Dose-response curve of each mouse OR mutant. (A) D3.39E mutants of Olfr339, Olfr1352, Olfr1353 and Olfr1377, (B) Y3.43L mutants of Olfr978 and Olfr979, (C) Olfr960 L3.39E, Y3.43L and double mutants, (D) ORs with reduced agonist response due to mutations. Olfr992D3.39E confronting octanol and Olfr62F3.43L against two-coumaranone. All constructs, including the vector control, were besides tested under RTP1S co-expression conditions. Fault bars indicate s.e.yard (n = 3).
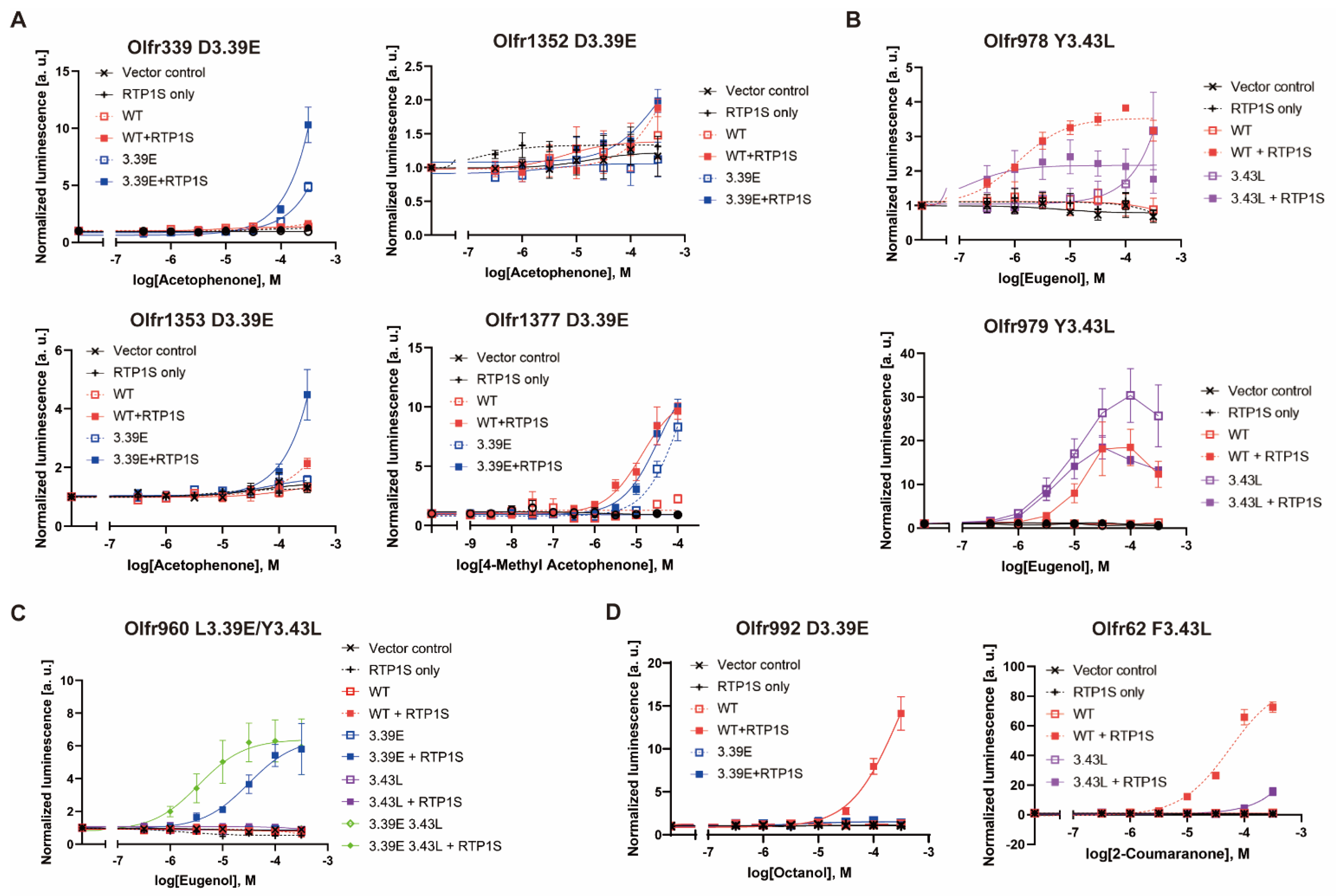
Figure 5. The alter in conserved E3.39 and L3.43 led to a loss of function. (Top) The cell-surface expression of the Rho-tagged wild-blazon, E3.39D and L3.43Y mutants of Olfr1508 and Olfr733. Each OR was transfected into HEK293T cells, and the cell surface expression level was measured. Ten-centrality: PE fluorescence, Y-axis: cell number. Left graph: Comparing between the WT and D3.39E mutant. The results from ii independent experiments are shown in the graph. Right: The effect of RTP1S co-expression on them. (Lesser) Dose-response curve of Olfr1508 and Olfr733 mutants to CuCl2 (Cu2+) and hexanal, respectively. Error bars indicate south.due east.m (northward = iii). Multiple comparisons were performed using one-mode ANOVA followed by Dunnett's test (** p < 0.01, *** p < 0.001).
Figure 5. The change in conserved E3.39 and L3.43 led to a loss of function. (Superlative) The prison cell-surface expression of the Rho-tagged wild-type, E3.39D and L3.43Y mutants of Olfr1508 and Olfr733. Each OR was transfected into HEK293T cells, and the cell surface expression level was measured. 10-axis: PE fluorescence, Y-axis: cell number. Left graph: Comparison between the WT and D3.39E mutant. The results from two independent experiments are shown in the graph. Right: The consequence of RTP1S co-expression on them. (Bottom) Dose-response curve of Olfr1508 and Olfr733 mutants to CuCl2 (Cu2+) and hexanal, respectively. Error confined indicate s.e.one thousand (n = iii). Multiple comparisons were performed using one-way ANOVA followed past Dunnett'southward test (** p < 0.01, *** p < 0.001).
Effigy 6. Furnishings of iii.39E and 3.43L mutants on human ORs (A,B) Phylogenic tree of mouse ORs showing the E3.39 or others, or L3.43 or others in 390 intact human ORs registered in the HORDE database. (C,D) Cell-surface expression of the Rho-tagged ORs and 3.39E mutants. Each mutant was transfected into HEK293T cells, and the jail cell surface expression level was measured. X-axis: PE fluorescence, Y-centrality: cell number. (E,F) Dose response curves of OR7D4 (WT and muant) and OR10G7 (WT and mutant) to androstenone and eugenol, respectively. Fault bars indicate southward.e.one thousand (n = 3). Multiple comparisons were performed using one-way ANOVA followed by Dunnett's test (* p < 0.05, ** p < 0.01, *** p < 0.001).
Figure 6. Effects of 3.39E and 3.43L mutants on human being ORs (A,B) Phylogenic tree of mouse ORs showing the E3.39 or others, or L3.43 or others in 390 intact human being ORs registered in the HORDE database. (C,D) Cell-surface expression of the Rho-tagged ORs and 3.39E mutants. Each mutant was transfected into HEK293T cells, and the cell surface expression level was measured. Ten-centrality: PE fluorescence, Y-centrality: cell number. (East,F) Dose response curves of OR7D4 (WT and muant) and OR10G7 (WT and mutant) to androstenone and eugenol, respectively. Mistake bars indicate southward.e.thousand (n = three). Multiple comparisons were performed using ane-style ANOVA followed by Dunnett'southward test (* p < 0.05, ** p < 0.01, *** p < 0.001).
| Publisher'south Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open up admission commodity distributed nether the terms and atmospheric condition of the Creative Eatables Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Source: https://www.mdpi.com/1422-0067/23/1/277/htm
 ,
,
Post a Comment for "Rtp Family Members Induce Functional Expression of Mammalian Odorant Receptors"